Ubiquitin-Proteasome_Dependent_Proteolysis
发布时间:2019-12-11 15:23 来源:SABiosciences
- 通路
- 概述
Review
The Proper functioning of a cell requires careful control of the levels of important structural proteins, enzymes, and regulatory proteins. Protein molecules are continuously synthesised and degraded in all living organisms. The concentration of individual cellular proteins is determined by a balance between the rates of synthesis and degradation, which in turn are controlled by a series of regulated biochemical mechanisms. Differences in the rates of protein synthesis and breakdown result in cellular and tissue atrophy (loss of proteins from cells) and hypertrophy (increase in protein content of cells). Precise control of protein turnover is, therefore, essential to cellular survival. The only way that cells can reduce the excessive level of a particular protein is by proteolytic degradation. Although protein breakdown can take place in the mitochondria, chloroplasts, the lumen of the endoplasmic reticulum and the endosomes, it occurs most commonly in one of two major sites of intracellular proteolysis: lysosomes, and the cytosol. Most non-selective protein degradation takes place in the lysosomes, whereas, short-lived regulatory proteins are degraded in the cytosol by intracellular proteolytic mechanisms. Two systems that play important roles in proteolysis in cytosol are the Calpain Proteases and the Ub (Ubiquitin)-Proteasome complex. The Ub-Proteasome complex, which consists of the Ub-conjugating system and the Proteasome, functions widely in intracellular protein turnover. It plays a central role in degradation of short-lived and regulatory proteins important in a variety of basic cellular processes. These complex degradation processes are controlled via specific degradation of a single or a subset of proteins (Ref.1, 2 & 3).
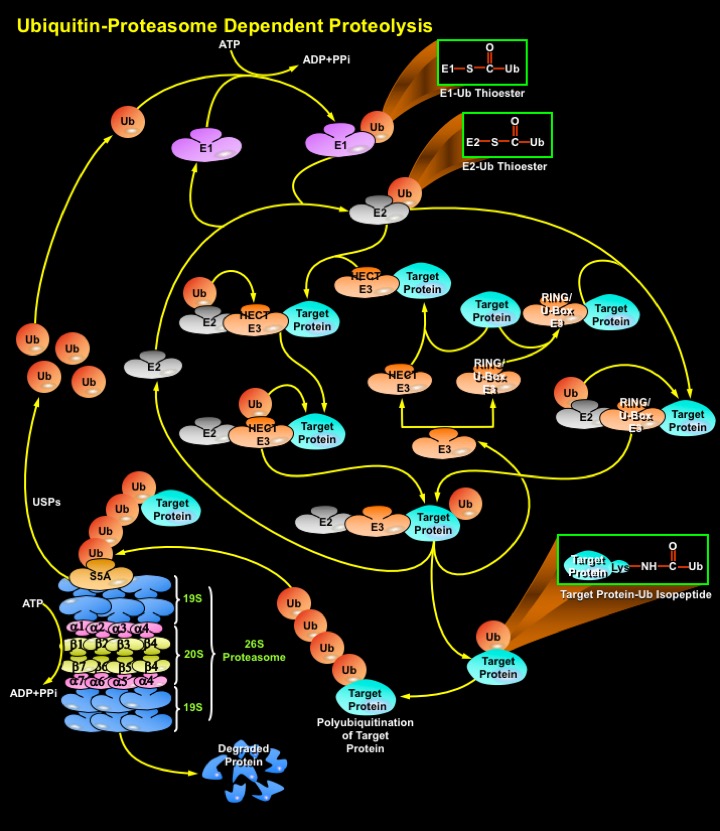
Degradation of a protein by the Ub-Proteasome pathway involves two distinct and successive steps: (i) covalent attachment of multiple Ub molecules to the target protein; and (ii) degradation of the tagged protein by the 26S Proteasome complex. Ub, a protein of 76 amino acids, binds covalently to available Lysine residues on target proteins, which are then recognised by Proteases. Ub undergoes an ATP-dependent reaction with proteins, which condenses its C-terminal Glycine residues with Lysine amino groups on the target protein. Such modified proteins are degraded soon afterward by the Proteasome. The Proteasome is a multicatalytic proteinase complex localized in the nucleus and cytosol. However, activity of the Ub-Proteasome system is not only limited to the cytosol and the nucleus, membrane-anchored and even secretory pathway-compartmentalized proteins are also targeted by the system. These proteins destined for degradation must be first translocated in a retrograde manner into the cytosol. To ensure efficient and specific removal of a certain protein at a certain time point, both Ub conjugation and degradation of the tagged substrates must be tightly regulated (Ref.4 & 5).
Proteins destined for degradation (Target Proteins) through the Ub-Proteasome pathway are first labeled by the Ub conjugation pathway. Ubiquitination is mediated by the sequential action of a multi-enzymatic system consisting of E1 (Ubiquitin-Activating Enzyme), E2/UBCs (Ubiquitin-Carrier Proteins or, Ubiquitin-Conjugating Enzymes), and E3 (Ubiquitin-Ligating Enzymes). An E1 activates Ub via the generation of a high energy thioester bond between the COOH-terminal Glycine on Ubiquitin and an internal E1 Cysteine residue in an ATP (Adenosine Triphosphate)-dependent manner. ATP is converted to ADP (Adenosine Monophosphate) and PPi (Inorganic Phosphate). Following activation, Ub is transferred in a second thioester linkage to one of several E2 enzymes. The E3s bind to the ubiquitinated E2s and to specific substrate proteins, thereby targeting the E2s to the substrate/target proteins and promoting the ligation of the Ub to the substrate, marking it for degradation by the Proteasome. Both E2 and E3 proteins exist as large families and different combinations of E2s with different E3 proteins define the substrate specificity. There are three distinct E3 families: the HECT (Homologous to the E6-AP Carboxyl Terminus), RING (Really Interesting New Gene) finger, U-box domain types, that differ in the mechanisms of transfer of the Ub to the substrate. In the case of the HECT domain-containing E3s, Ub is first transferred from the thioester linkage on the E2 to a thioester linkage on the E3, and then to the target. In contrast, RING-finger and U-box E3s do not have a direct catalytic role in protein ubiquitylation but act as facilitators of interaction' between an E2 and target protein. These Ub ligases do not form thioester bonds with the Ub, but rather promote the transfer of Ub directly from the E2 to the final substrate. RING finger and U-box domains are structurally related. Thus, all the three types of E3s mediate the covalent attachment of multiple Ub moieties to the target protein. The first moiety is transferred to an NH2 group of a Lysine residue of the protein substrate to generate an Isopeptide bond. In successive reactions, a Polyubiquitin chain is synthesized by transfer of additional Ub moieties to the previously conjugated Ub. The chain serves as a recognition marker for the Proteasome, and directs the target protein to the 26S Proteasome complex for degradation (Ref.2, 6, 7 & 8).
The proteolytic activities of the 26S Proteasome are located within a 20S multisubunit structure, that contains multiple Peptidase activities and functions as the catalytic machine. It is composed of 28 subunits arranged in four heptameric, tightly stacked, rings (Alpha7, Beta7, Beta7, Alpha7) to form a cylindrical structure. The Alpha-subunits make up the two outer and the Beta-subunits the two inner rings of the stack. The entrance of target proteins to the active site of the complex is guarded by the Alpha-subunits that allow access only to unfolded and extended polypeptides. The proteolytic activity is confined to the Beta-subunits. When capped by the 19S regulatory complex at each end, the 20S complex forms the core of the 26S proteasome. Once Ub-tagged, proteins bind to S5A (Protease 26S Subunit-5A) subunits in the 19S regulatory cap of the Proteasome, where they are deubiquitinated and unfolded in an energy-dependent manner. Isopeptidases/USPs (Ubiquitin-Specific Proteases) cleave off the Ubiquitin moieties, which are recycled, and the target protein enters the catalytic inner chamber of the Proteasome, the 20S complex. Peptidases on the inner surface of Beta-subunits degrade the substrate, releasing Ubiquitinated peptides of 3–22 amino acids in size, which exit the Proteasome. Proteasomes use ATP to drive conformational changes in their subunits. ATP hydrolysis is not needed to actually cleave the peptide bonds of a protein, but instead is thought to involve recognition of target proteins, their unfolding, translocation of the target protein into the Proteasome's chamber, and/or the opening and closing of Proteasome gates. The system has high specificity and selectivity. Since the target proteins bind to the ligases prior to conjugation, E3s are key players in determining the specificity of the system, and they represent a class of "drugable" targets for pharmaceutical intervention. These E3 ligases or their components are overexpressed in many human Cancers and their inhibition leads to growth suppression or apoptosis (Ref.8, 9 & 10).
With the multiple cellular targets, the Ub-Proteasome system is involved in the regulation of many basic cellular processes such as cell cycle, differentiation and development, the response to stress and extracellular modulators, morphogenesis of neuronal networks, modulation of cell surface receptors and signal transduction pathways, ion channels and the secretory pathway, DNA repair, regulation of the immune and inflammatory responses, antigen processing and presentation, biogenesis of organelles, apoptosis and in maintaining the integrity of the proper folded state of proteins. These intracellular proteolytic systems recognize and destroy misfolded or damaged proteins, unassembled polypeptide chains, short-lived regulatory proteins and abnormal proteins that result from oxidative stress and mutations, which might otherwise disrupt normal cellular homeostasis. Considering the broad range of substrates and processes in which the Ub-Proteasome pathway is involved, aberrations in the system have been implicated in the pathogenesis of several diseases, both inherited and acquired. The pathological states can be divided into two groups: (i) those that result from loss of function, a mutation in an enzyme or substrate that leads to stabilization of certain proteins; and (ii) those that result from a gain of function, resulting in accelerated degradation (Ref.11 & 12).
References
- 1
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome.
- 2
- Wilkinson KD, Ventii KH, Friedrich KL, Mullally JE. The ubiquitin signal: assembly, recognition and termination. Symposium on ubiquitin and signaling.
- 3
- Ciechanover A, Iwai K. The ubiquitin system: from basic mechanisms to the patient bed.
- 4
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction.
- 5
- Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction.
- 6
- Robinson PA, Ardley HC. Ubiquitin-protein ligases.
- 7
- Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation.
- 8
- Kierszenbaum AL. The 26S proteasome: ubiquitin-mediated proteolysis in the tunnel.
- 9
- Hartmann-Petersen R, Seeger M, Gordon C. Transferring substrates to the 26S proteasome.
- 10
- Sun Y. Targeting E3 ubiquitin ligases for cancer therapy.
 关于我们
关于我们