Transcription_of_mRNA
发布时间:2019-12-11 15:06 来源:SABiosciences
- 通路
- 概述
Review
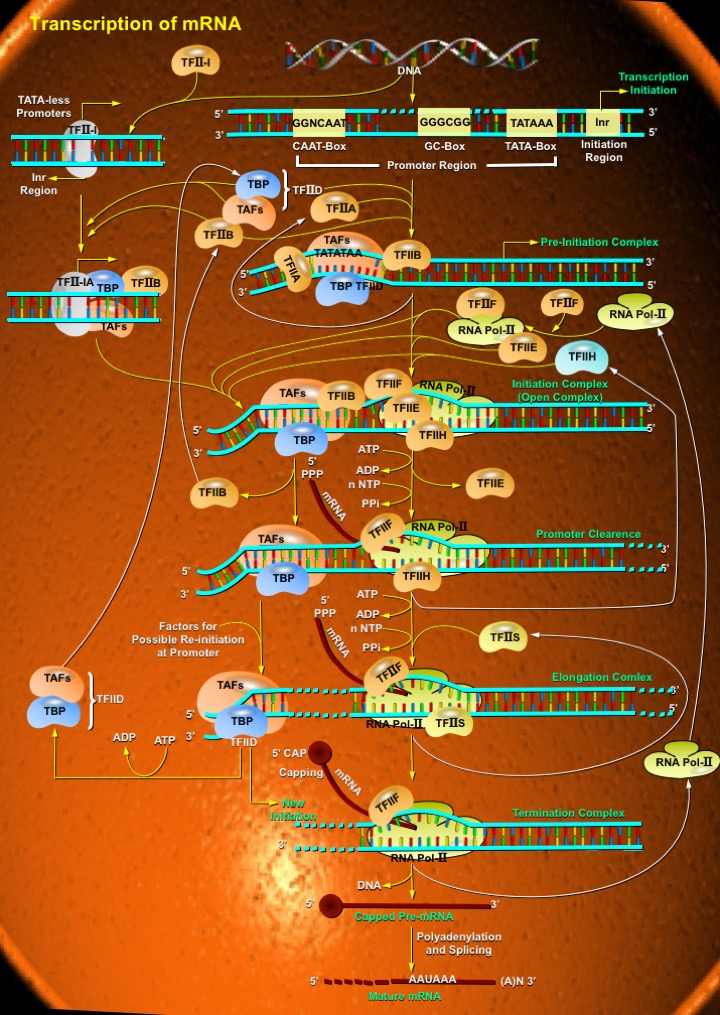
Transcription is the process through which a DNA sequence is enzymatically copied by an RNA polymerase to produce a complementary RNA. Transcription can also be defined as a process that transcribes genetic information from DNA into RNA. In eukaryotes, it takes place in the nucleus, mitochondria and chloroplast. Transcription is performed by DNA-directed RNA Polymerases. Unlike DNA Polymerases, RNA Polymerases do not need a primer to start the reaction. While Bacteria contain only 1 RNA Polymerase, there are 3 different RNA polymerases in eukaryotic cells, which catalyzes the synthesis of three types of RNA. RNA Pol-I (RNA Polymerase-I) is located in the nucleolus and transcribes rRNA (ribosomal RNA). RNA Pol-II (RNA Polymerase-II) is localized to the nucleus, and transcribes mRNA (messenger RNA) and most snRNAs (small nuclear RNAs). RNA Pol-III (RNA Polymerase-III) is localized to the nucleus (and possibly the nucleolar-nucleoplasm interface), and transcribes tRNA (transfer RNA) and other small RNAs (including the small 5S rRNA). All these polymerases are multisubunit complexes: two large polypeptides (200 and 140 kDa) are associated with about 12 smaller subunits, some of which are common to all three enzymes. Additionally there are different Pol in mitochondria and chloroplast. mRNA is the RNA that encodes and carries information from DNA during transcription to sites of protein synthesis to undergo translation in order to yield a gene product. In most mammalian cells, only 1% of the DNA sequence is copied into a functional mRNA. Only one part of the DNA is transcribed to produce nuclear RNA, and only a minor portion of the nuclear RNA survives the RNA processing steps. Like DNA replication, mRNA transcription proceeds in the 5' ¡ú 3' direction (ie the old polymer is read in the 3' ¡ú 5' direction and the new, complementary fragments are generated in the 5' ¡ú 3' direction). Transcription is divided into 3 stages: Initiation, Elongation and Termination (Ref.1 & 2).
Initiation is the first step in mRNA transcription. It includes the construction of the RNA Polymerase Complex on the gene's promoter with the help of transcription factors. In order to start transcription, a core promoter element at the DNA, e.g. the TATA box must be recognized and bound by TBP (TATA Binding Protein) subunit of the basic transcription factor TFIID (Transcription Factor for Polymerase-IID), introducing sharp kinks to the DNA. Besides TBP, TFIID also contains more than 8 other subunits known as TAFs (TATA box binding protein (TBP)-Associated Factors). The sequence TATA is located ¡®-30¡¯ nucleotides upstream of the TSP (Transcription Start Point). In addition, there are also some weakly conserved features including the BRE (B-Recognition Element), approximately 5 nucleotides upstream of the TATA Box. TFIIB (Transcription Factor for Polymerase-IIB) and TFIIA (Transcription Factor for Polymerase-IIA) stabilize this interaction between TATA and TFIID, forming a Pre-Initiation Complex. Frequently, these steps are controlled by activation or repression mechanisms. In case of promoters not containing the TATA-sequence, the Inr (Initiator) motif of DNA can mediate initiation. It is recognized by Inr-binding proteins (such as TFII-I to which TFIID associates) or by subunits TAFII250 and TAFII150 of TFIID. This commonly occurs with house keeping genes. RNA Pol-II together with transcription factors TFIIE (Transcription Factor for Polymerase-IIE), TFIIF (Transcription Factor for Polymerase-IIF), and TFIIH (Transcription Factor for Polymerase-IIH) is then recruited to form the Initiation Complex. TFIIF and RNA Pol-II (two subunits, RAP30 and RAP74, showing some similarity to bacterial sigma factors) enter the complex together. TFIIF helps to speed up the polymerization process. TFIIE enters the complex, and helps to open and close the PolII¡¯s Jaw like structure, which enables movement down the DNA strand. TFIIE and TFIIH enter concomitantly. TFIIH is a large protein complex that contains among others the CDK7 (Cyclin-Dependent Kinase-7)/Cyclin-H Kinase complex and a DNA Helicase. TFIIH has three functions: it binds specifically to the template strand to ensure that the correct strand of DNA is transcribed and melts or unwinds the DNA (ATP dependently) to separate the two strands using its Helicase activity. It has a kinase activity that phosphorylates the CTD (C-terminal domain) of Pol II at the amino acid serine. This switches the RNA polymerase to start producing RNA, which marks the end of initiation and the start of elongation. Finally it is essential for NER (Nucleotide Excision Repair) of damaged DNA. TFIIH and TFIIE strongly interact with one another. TFIIE affects TFIIH¡¯s catalytic activity. Without TFIIE, TFIIH will not unwind the promoter. TFIIB, assisted by TFIIF, acts as a bridge to PolII. This is rate limiting step. At this stage the DNA strands start to become separated in an ATP requiring reaction forming an Open initiation complex (Ref. 3 & 4).
Transcriptional Initiation is regulated by many mechanisms. These can be separated into two main categories: Protein interference and Chromatin structure inhibition. Protein interference is the process where some signaling protein interacts, either with the promoter or some stage of the partially constructed complex, to prevent further construction of the Polymerase complex, so preventing Initiation. This is generally a very rapid response and is used for fine level, individual gene control and for cascade processes for a group of genes useful under a specific conditions (for example DNA repair genes or heat shock genes). Chromatin structure inhibition is the process where the promoter is hidden by chromatin structure. Chromatin structure is controlled by post-transcriptional modification of the Histones involved and leads to gross levels of high or low transcription levels. These methods of control combined in a modular method, allow very high specificity in transcription initiation control (Ref. 5 & 6).
The second step in the process of Transcription is Elongation. This step involves the actual transcription of the majority of the gene into a corresponding RNA sequence, highly moderated by several methods. The initiation complex becomes active when PolII transcribes the first few bases close to the promoter, beginning at the transcription start site. The maximum length of the RNA-DNA hybrid is only 2¡ª3 bp. RNA chain elongation involves a series of forward movements interspersed by pauses. The transition from initiation to elongation (promoter clearance by PolII) is still poorly defined. For catalyzing elongation, PolII gets highly phosphorylated at the CTD (Carboxy Terminal Domain) of the largest subunit, which causes a confirmation change and subsequent clearance from TFIID Complex. This reaction is catalyzed by TFIIH and TFIIE. The CTD domain contains a tandemly repeated heptapeptide (YSPTSPS) n (one letter code for amino acids, n=52 in mammals). Upon clearing from the TFIID-complex, Topoisomerase I presumably moves to the elongation complex and facilitates elongation. Also, TFIIH exerts its ATP dependent helicase activity, after TFIIE (which inhibits this activity) has left the complex. Highly supercoiled DNA does not require this activity, possibly due to its inherent energy content. Purified PolII alone only transcribes 1.5----5nt/sec. By action of elongation factors (TFIIA, TFIIF and Elongin/SIII), transcription rates in vivo increase to 20¡33nt/sec. RNA Pol-II elongation promoters are divided in 3 classes: Drug/sequence-dependent arrest affected factors (Eg. SII (TFIIS) and PTEF-b protein families); Chromatin structure oriented factors (phosphorylation, acetylation, methylation and ubiquination) and RNA Pol-II catalysis improving factors, which improve the Vmax or Km of RNA Pol-II, so improving the catalytic quality of the Polymerase enzyme (Eg. TFIIF, Elongin and ELL families). Elongation downregulation is also possible, in this case usually by blocking Polymerase progress or by deactivating the Polymerase (Ref. 3, 7 & 8).
The final step in mRNA transcription is Termination i.e, the cessation of RNA transcription and the disassembly of the RNA Polymerase complex. The transcriptional machinery continues 0.5 to 2 kb beyond of the Poly (A) signal and then dissociates. The exact mechanism is unknown. In eukaryotes using RNA Pol-II this termination is very variable (up to 2000 bases), relying on post transcriptional modification. During the course of transcription, the initial RNA product synthesized by RNA Pol-II, called a Primary transcript undergoes several processing steps including Capping, Splicing and Polyadenylation, before a functional mRNA is produced. Shortly after transcription begins, the 5' end of the nascent RNA is capped with 7-Methyl-Guanylate.Transcription by RNA Pol-II terminates at any one of the multiple sites approx. 0.5-2kb downstream from the 3' end of the last exon in the transcript. The 3' end of a functional mRNA then is generated by endonucleolytic cleavage at a specific sequence, the Poly-A site, located at the 3' end of the final exon. A stretch of 100-250 Adenine residues is added to the 3'- Hydroxyl group left by the cleavage reaction. Finally, introns are removed by RNA Splicing before the completed RNA is transported to the cytoplasm (Ref.1, 9 & 10).
References
- 1
- Taylor WR. Transcription and translation in an RNA world.
- 2
- Batey RT. Structures of regulatory elements in mRNAs.
- 3
- Deng W, Roberts SG. Core promoter elements recognized by transcription factor IIB.
- 4
- Cramer P. Recent structural studies of RNA polymerases II and III.
- 5
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD.
- 6
- Juven-Gershon T, Hsu JY, Kadonaga JT. Perspectives on the RNA polymerase II core promoter.
- 7
- Reyes-Reyes M, Hampsey M. A role for the Ssu72 CTD phosphatase in RNA polymerase II transcription elongation.
- 8
- Wright KJ, Marr MT 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter.
- 9
- Geiduschek EP, Kassavetis GA. Transcription: adjusting to adversity by regulating RNA polymerase.
- 10
- Galamb O, Sipos F, Dinya E, Spisak S, Tulassay Z, Molnar B. mRNA expression, functional profiling and multivariate classification of colon biopsy specimen by cDNA overall glass microarray.
 关于我们
关于我们