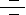RNAi_Pathway
发布时间:2019-12-11 12:01 来源:SABiosciences
- 通路
- 概述
Review
RNAi (RNA interference) is a cellular pathway of gene silencing in a sequence-specific manner at the mRNA (messenger RNA) level. The basic mechanism behind RNAi is the breaking of a dsRNA (double-stranded RNA) matching a specific gene sequence into short pieces of RNAs called siRNA (small interfering RNA). siRNAs are 21–23nt (nucleotide) dsRNA duplexes with symmetric 2–3nt 3' overhangs and 5'-phosphate and 3'-hydroxyl groups, which trigger the degradation of mRNA matching its sequence (Ref.1). Interference of gene expression by siRNA is now recognized as a naturally occurring biological strategy for silencing alleles during development in plants, invertebrates, and vertebrates.
RNA silencing occurs in a broad range of eukaryotic organisms including fungi (Quelling), animals (RNAi), and plants (PTGS, Post-Transcriptional Gene Silencing). In all these organisms, the process is triggered by dsRNA and requires a conserved set of gene products. The mechanism for RNA silencing involves the chopping of dsRNA into smaller pieces of a defined length corresponding to both sense and antisense strands of the target gene by the Rnase-III (Ribonuclease-III) family member, Dicer in an ATP-dependent reaction. Dicer chops dsRNA into two classes of smaller RNAs—miRNAs (microRNAs) and siRNAs—that are around 21–23nts in length. Dicer delivers the siRNAs to a group of proteins called the RISC (RNA-Inducing Silencing Complex), which uses the antisense strand of the siRNA to bind to and degrade the corresponding mRNA, resulting in gene silencing (Ref.2). There is a strict requirement for the siRNA to be 5' phosphorylated to enter into RISC, and siRNAs that lack a 5' phosphate are rapidly phosphorylated by an endogenous kinase. Although the uptake of siRNAs by RISC is independent of ATP, the unwinding of the siRNA duplex requires ATP. Once unwound, the single-stranded antisense strand guides RISC to mRNA that has a complementary sequence, which results in the endonucleolytic cleavage of the target mRNA. The target mRNA is cleaved at a single site in the centre of the duplex region between the guide siRNA and the target mRNA, 10nt from the 5' end of the siRNA for degradation (Ref.3).
Endogenously expressed siRNAs have not been found in mammals. However, the related short miRNAs are produced by Dicer cleavage, which causes translational repression. miRNAs are expressed polycistronically and are processed through at least two sequential steps: (i) generation of longer (~70nt) endogenous precursors with imperfect hairpin RNA structures from the longer transcripts (termed pri-miRNAs); and (ii) processing of pre-miRNAs into mature miRNAs. Both the steps are compartmentalized into the nucleus and cytoplasm, respectively, and the pre-miRNA serves as the substrate for nuclear export (Ref.6). The identities of RNA Polymerase and the nuclear processing enzyme have not been determined yet. The miRNAs bind to sites that have partial sequence complementarity in the 3' UTR (Untranslated Region) of the target mRNA, causing repression of translation and inhibition of protein synthesis. In addition to Dicer, other PPD (PAZ/PIWI Domain Proteins), including eIF2C2 (Eukaryotic Translation Initiation Factor-2C2) also function in both pathways (Ref.3). Similar complexes also target chromatin remodeling in the nucleus.
RNA silencing, triggered by dsRNA molecules, is now recognized as a mechanism for cellular protection and cleansing. It defends the genome against molecular parasites such as viruses and transposons, while removing abundant but aberrant nonfunctional mRNAs (Ref.4). However, the applicability of this approach is limited in mammals because the introduction of dsRNA longer than 30nt induces a sequence-nonspecific interferon response. Interferon triggers the degradation of mRNA by inducing 2'-5' oligoadenylate synthase. In addition, interferon activates PKR (Protein Kinase-R), which phosphorylates eIF2-Alpha leading to global inhibition of mRNA translation (Ref.3). siRNAs can also suppress HIV1 (Human Immunodeficiency Virus Type-1) replication in cell lines and proliferating CD4 T-Cells. The possibility of stable expression of siRNAs may pave the road for new gene therapy applications, such as treatment of persistent viral infections (Ref.5).
References
- 1
- Cioca DP, Aoki Y, Kiyosawa K. RNA interference is a functional pathway with therapeutic potential in human myeloid leukemia cell lines.
- 2
- Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells.
- 3
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression.
- 4
- Tijsterman M, Ketting RF, Plasterk RH. The genetics of RNA silencing.
- 5
- Tuschl T. Expanding small RNA interference.
- 6
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization.
 关于我们
关于我们