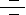Notch_Signaling
发布时间:2019-12-11 10:57 来源:SABiosciences
- 通路
- 概述
Review
The Notch signaling pathway is a fundamental signaling system used by neighboring cells to communicate with each other in order to assume their proper developmental role. Notch proteins are cell surface transmembrane-spanning receptors which mediate critically important cellular functions through direct cell-cell contact. Interaction between Notch and its proposed ligands initiates a signaling cascade that governs cell fate decisions such as differentiation, proliferation, and apoptosis in numerous tissue types. The core elements of the Notch signaling system include the Notch receptor, DSL ligands (Delta and Serrate/Jagged in Drosophila and vertebrates, Lag2 in Caenorhabditis elegans) and CSL DNA-binding proteins (CBF1/RBPJ-kappa in vertebrates, Su(H) [Suppressor of hairless] in Drosophila, Lag1 in C. elegans). Four paralogs of the Notch gene, Notch1, 2, 3 and 4, and five Notch ligands, including Jagged1, Jagged2, Delta1, Delta2 and Delta3, have been identified in vertebrates (Ref.1). The growing array of multiple genes for each ligand as well as genes for numerous modulators of Notch pathway confers extensive complexity on this signaling system. (Ref.2).
Notch proteins (and ligands) contain extracellular EGF (Epidermal Growth Factor)-like repeats, which interact with the DSL domain of ligands. Activation of Notch upon ligand binding is accompanied by proteolytic processing that releases an intracellular domain of Notch (NICD) from the membrane tether. The NICD contains the RAM23 domain (RAM), which enhances interaction with CSL protein, NLS (Nuclear Localization Signals), a CDC10/Ankyrin repeat domain ANK, which mediates interactions with CSL and other proteins, and a PEST domain rich in proline, glutamate, serine and threonine residues (Ref.6). The Notch COOH-terminal fragment NEXT is cleaved by Gamma-secretase (includes Presenilin and Nicastrin) to release NICD into the cytoplasm. Upon release, the NICD translocates to the nucleus and associates with the CSL [CBF1/RBPJ-kappa/Su (H)/Lag1] family of DNA-binding proteins to form a transcriptional activator, which activate the expression of a set of target genes, including the E (spl) (Enhancer of Split) group and others (Ref.1). During its activation, Notch is cleaved at least three times. First, as part of the biosynthetic processing of Notch, a Furin-like protease in the Golgi converts nascent Notch proteins into heterodimers. This "S1 cleavage" is necessary for cell-surface expression of Notch but is not directly involved in the ligand-induced release of the active intracellular domain. Instead, ligands binding to the Notch heterodimer trigger a concerted proteolysis at two additional sites S2 and S3. The S2 cleavage severs most of the Notch extracellular domain, and S3 cleavage, which occurs within the transmembrane domain, releases the transcriptionally active intracellular domain (Ref.3).
Most of the Notch target genes encode transcription regulators, which in turn modulate cell fate by affecting the function of tissue-specific basic helix-loop-helix HES gene family (mammalian homologues of Drosophila Hairy and E (spl) such as HES1 and HES5) or through other molecular targets, such as NF-kappaB (Ref.4). These in turn regulate expression of tissue-specific transcription factors that influence lineage commitment and other events. Other potential Notch targets include p21/WAF1 (Wild type p53-Activated Fragment-1)/CIP1 (Cyclin-Dependent Kinase Inhibitor-1), Cyclin-D1, HERP, and MAPK (Mitogen-Activated Protein Kinase) phosphatase LIP1 (Ref.5). Activation of Notch by DSL ligands proteolytically releases the Notch intracellular domain from the plasma membrane, and the resulting protein directly translocates to the nucleus to participate in the transcriptional regulation of target genes (Ref.3). Notch may act through two pathways: (i) as a transcription factor to regulate gene expression at the level of transcription and (ii) as a stimulator of protein turnover through a mechanism involving the proteasome-mediated destruction of Notch-targeted substrates. Once in the nucleus, NICD converts CSL from a transcriptional repressor to a transcriptional activator. This conversion occurs by direct protein-protein interactions between the NICD, SKIP (Ski-Related Protein) and CSL, which leads to SMRT (Silencing Mediator of Retinoid and Thyroid Hormone Receptor)/HDACs (Histone Deacetylases) dissociation. Notch/CSL recruit HATs (Histone Acetylases) to assist in chromatin remodeling, and Mastermind/Lag3 to activate additional targets. The metabolism of NICD in the nucleus is controlled by phosphorylation and ubiquitination by the E3 Ubiquitin Ligase Sel10 and Su (Dx) (Suppressor of Deltex). NICD degradation resets the cell and prepares it for the next round of Notch signaling (Ref.6). In this newly recognized role, Notch acts to prevent cells from acquiring neural or myogenic competence earlier in development. This activity requires Deltex (Dx), a cytoplasmic ring finger protein and the kinase GSK3Beta (Sgg). Several additional factors that influence signaling include the ligand Serrate and its negative regulator Fng (Fringe); the metalloproteases TACE (also known as ADAM17), Kuz (Kuzbanian), which acts as a Delta and potentially as a Notch-processing enzyme; the trans-Golgi convertase Furin, which cleaves Notch; Presenilin, which may cleave Notch in the membrane; the NICD interacting proteins Dsh (Disheveled), Dab (Disabled), and Numb; and in the nucleus, the regulator Hairless (H) (Ref.8). The LNR (Lin/Notch Repeat) domain maintains the association between the polypeptides resulting from the Furin cleavage (Ref.6). WNT acts to block Notch by stimulating Dsh to inhibit GSK3Beta (Glycogen Synthase Kinase-3Beta) activity or to circumvent Deltex/Notch interaction (Ref.6).
The Notch signaling pathway is an evolutionarily conserved, intercellular signaling mechanism essential for proper embryonic development in organisms as diverse as insects, nematodes, echinoderms and mammals (Ref.7). Notch receptors initiate a highly conserved signaling pathway that influences cell fate decisions within multiple tissues and regulate the ability of precursor cells to respond to other developmental signals. In mammals, Notch signaling regulates neurogenesis, myogenesis, vasculogenesis, hematopoiesis, skin development, and other aspects of organogenesis. In addition, Notch signaling is involved in other critical cellular processes such as proliferation and apoptosis. Consistent with the ability to influence cellular differentiation in multiple tissues, mutations of Notch receptors and components of its signaling pathway have been associated with a number of diseases, including human T-Cell leukemia (Notch1), CADASIL (Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy) (Notch3), Spondylocostal Dysostosis, and Alagille syndrome (Jagged1). Three proteins essential for Epstein-Barr virus transformation of B-Cells—EBNA2, EBNA3a, and EBNA3c—each of which bind to CSL and modifies Notch activity, also directly target the Notch pathway. Also, the murine Notch4 gene has been identified as an integration site of Mammary Tumor Virus, resulting in constitutive activation of Notch4 and breast carcinoma (Ref.5). During tooth development, Notch signaling is associated with the differentiation of dental epithelial and mesenchymal cells and is also involved in the regulation of the stem cells in the continuously growing incisor (Ref.7). Appropriate manipulation of Notch signaling may become a useful tool in addressing a variety of human dysplastic conditions as well as tissue regeneration (Ref.8).
References
- 1
- Ohishi K, Varnum-Finney B, Flowers D, Anasetti C, Myerson D, Bernstein ID. Monocytes express high amounts of Notch and undergo cytokine specific apoptosis following interaction with the Notch ligand, Delta-1.
- 2
- Jeffries S, Capobianco AJ. Neoplastic transformation by Notch requires nuclear localization.
- 3
- Kramer H. RIPping notch apart: a new role for endocytosis in signal transduction?
- 4
- Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, Velazquez OC, Herlyn M. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis.
- 5
- Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors.
- 6
- Kopan R. Notch: a membrane-bound transcription factor.
- 7
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling.
- 8
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development.
 关于我们
关于我们