Mismatch_Repair
发布时间:2019-12-11 10:29 来源:SABiosciences
- 通路
- 概述
Review
In all organisms, DNA repair processes are crucial to maintain the integrity of the genome through the generations. Many animals minimize the number of cell divisions and thus the chance to accumulate mutations from one to the next generation. The DNA MMR (Mismatch Repair) apparatus has an important function in stabilizing the genome, and the protein components of this system are highly conserved in all pro- and eukaryotic systems. The MMR system is responsible for the post-replicative repair of mismatches and small single stranded DNA loops, and it is critically involved in preventing recombination between homologous DNA sequences (Ref.1).
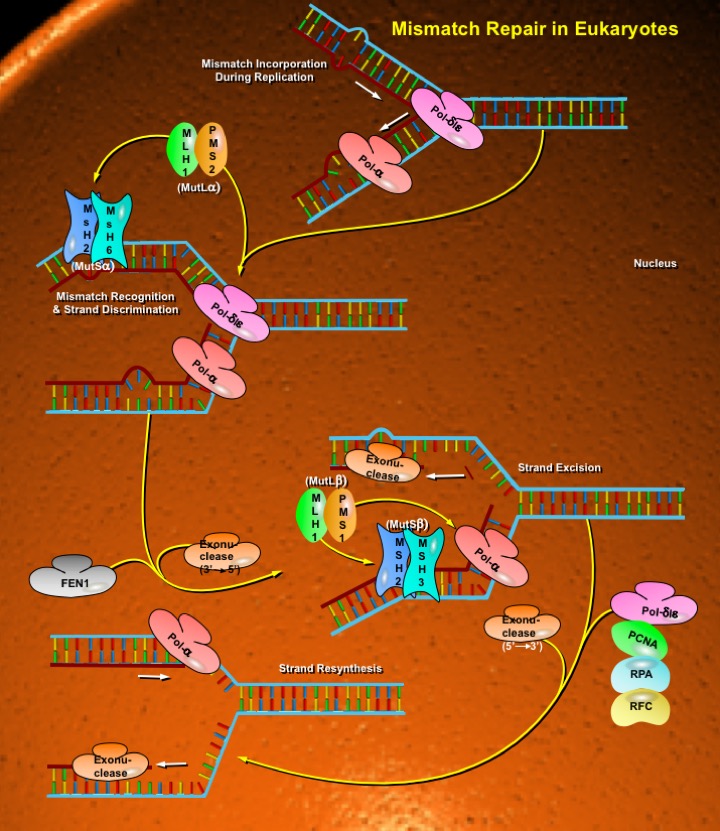
MMR removes nucleotides mispaired by DNA Polymerases and IDLs (Insertion/Deletion Loops) ranging from one to ten or more bases that result from slippage during replication of repetitive sequences or during recombination. Defects in this system dramatically increase mutation rates, fuelling the process of oncogenesis. Four principal steps involved in MMR are: (1) recognition of base-base mismatches and IDLs; (2) recruitment of additional MMR factors; (3) search for a signal that identifies the wrong (newly synthesized) strand, followed by degradation past the mismatch; and (4) resynthesis of the excised tract (Ref.2). There are several mispair-recognizing proteins. The eukaryotic MMR system contains proteins related to the bacterial MutS and MutL proteins but is more complex than the bacterial system. It involves two different heterodimeric complexes of MutS-related proteins, MSH2-MSH3 (known as MutSBeta) and MSH2-MSH6 (known as MutSAlpha), and each has different mispair recognition specificity. Heterodimers MSH2-MSH6 focuses on mismatches and single-base loops, whereas MSH2-MSH3 dimmers (MutSBeta) recognize ILDs. Similarly, instead of a single MutL-related protein, eukaryotic MMR involves a heterodimeric complex of two MutL-related proteins, MLH1-PMS1 (PMS2 in humans) (Ref.3). Heterodimeric complexes of MLH1/PMS2 (MutLAlpha) and MLH1/PMS1 (MutLBeta) interact with MSH complexes and replication factors. Excision and resynthesis of the nascent strand (containing the mismatch or IDL) is performed by a number of proteins including PCNA (Proliferating Cell Nuclear Antigen), RPA (Replication Protein-A), RFC (Replication Factor-C) Exonuclease-I, DNA Polymerases Delta/Epsilon, Endonuclease FEN1 (Flap structure-specific Endonuclease-1), and additional factors. MMR components also interact functionally with NER (Nucleotide Excision Repair) and recombination.
MMR has an important role in genetic recombination beyond the repair of mispaired bases in recombination intermediates. MMR regulate the extent of formation of heteroduplex tracts during recombination (Ref.5) by regulating the resolution of Holliday junctions. It also suppresses recombination between divergent sequences, a process that is similar to the regulation of heteroduplex tract formation. The MSH2 and MSH3 proteins also act in recombination between duplicated DNA sequences and have been implicated in the removal of nonhomologous DNA strands greater than 30 bases long at the ends of recombining segments. This reaction involves the NER (Nucleotide Excision Repair) complex Rad1-Rad10 (Ref.4). MMR is involved in cancer especially sporadic (non-familial) colorectal tumors that exhibit widespread alterations of poly (A) tracts. Some heritable mutations in the human MSH2 gene predispose patients to a cancer known as HNPCC (Hereditary Non-Polyposis Colorectal Carcinoma) (Ref.3).
References
- 1
- Jean M, Pelletier J, Hilpert M, Belzile F, Kunze R. Isolation and characterization of AtMLH1, a MutL homologue from Arabidopsis thaliana.
- 2
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer.
- 3
- Kolodner RD. Guarding against mutation.
- 4
- Bertrand P, Tishkoff DX, Filosi N, Dasgupta R, Kolodner RD. Physical interaction between components of DNA mismatch repair and nucleotide excision repair.
- 5
- Detloff P, White MA, Petes TD. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae.
 关于我们
关于我们