Factors_Promoting_Cardiogenesis_in_Vertebrate
发布时间:2019-12-10 11:03 来源:SABiosciences
- 通路
- 概述
Review
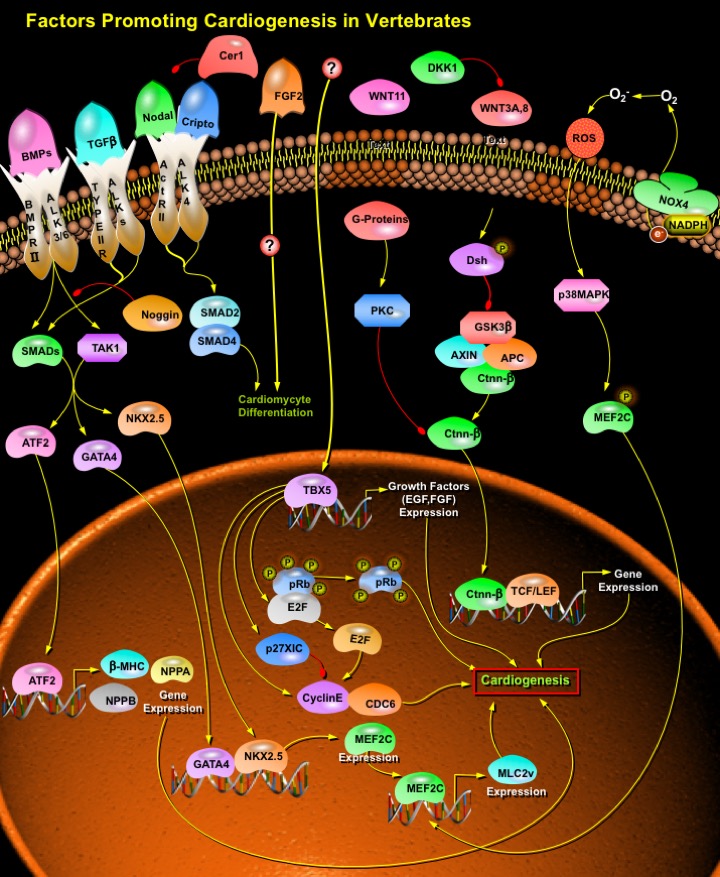
Heart is the first organ to form and function in the Embryo, and all subsequent events in the life of the organism depend on the Heart's ability to match its output with the organism's demands for Oxygen and nutrients. Abnormalities in Heart formation, the most common form of Human birth defects, affect nearly 1% of newborns, and their frequency in spontaneously aborted pregnancies is estimated to be tenfold higher. Heart development is an elaborate process requiring Cell specification, Cell differentiation, Cell migration, morphogenesis, and interactions among cells from several embryonic origins. The Heart is formed through multiple developmental steps, which include the determination of the Cardiac field in the mesoderm, differentiation of Cardiac precursor cells, and maturation of the Heart. Entry of cells into the Cardiac lineage is dependent upon appropriate external signals coupled to the expression of a set of transcription factors that initiates the program for Cardiac gene expression and drives the morphogenic events involved in formation of the multichambered Heart. Research in Mice, Birds, Amphibians, Flies and Mammals, as well as in various cell culture systems, has led to the identification of multiple transcription factors and extracellular growth factors whose concerted actions specify the Cardiac lineage in mesodermal progenitor cells. The earliest expressed transcription factors that initiate Cardiac fate are the homeobox transcription factor NKX2.5 (NK2 Transcription Factor Related Locus-5) and members of the GATA (GATA Binding Protein) family of zinc finger transcription factors, GATA4 (GATA Binding Protein-4), GATA5 (GATA Binding Protein-5), and GATA6 (GATA Binding Protein-6). Equally important roles in Heart development have been shown for members of the T-box (Tbx5 (T-Box-5), Tbx20 (T-Box-20)), basic helix-loop-helix (dHAND/HAND2 (Heart and Neural crest Derivatives expressed-2), eHAND/HAND1 (Heart and Neural crest Derivatives expressed-1)), and MADS (MCMI, Agamous, Deficiens, Serum response factor) domain (MEF2) families. LIM (a cysteine-rich motif identified in the homeobox genes lin-11, Isl-1, and mec-3) homeodomain transcription factor, which is expressed in a distinct population of Cardiac precursor cells, is essential for the formation of the outflow tract and the right ventricle. Extracellular signals that act upstream of these factors have been primarily identified by their ability to induce Cardiac differentiation in non-cardiac mesoderm. These signals belong to the BMP (Bone Morphogenetic Protein), FGF (Fibroblast Growth Factor), and WNT (Wingless-related MMTV integration site) families of Growth Factors and also include secreted WNT antagonists such as Dkk1 (Dickkopf1) and Crescent (Ref.1 & 2).
FGF2 (Fibroblast Growth Factor-2) and BMPs play a crucial role in early Cardiomyogenesis. FGF2 is required for the expression of Cardiac transcription factors and the differentiation of mesoderm explants induced by BMP2 (Bone Morphogenetic Protein-2). FGF2 induces mesenchymal cell formation from precardiac mesoderm explants. Other members of the FGF family compensate for the lack of FGF2 expression in the embryo, for instance FGF4 (Fibroblast Growth Factor-4) or FGF8 (Fibroblast Growth Factor-8). BMPs play a vital role in Cardiac development. BMP Receptors are essential for myocyte-dependent functions and signals in Cardiac organogenesis. BMPs like BMP2, BMP4 (Bone Morphogenetic Protein-4) and BMP5 (Bone Morphogenetic Protein-5), BMP7 (Bone Morphogenetic Protein-7), BMP10 (Bone Morphogenetic Protein-10), bind to Serine/threonine kinase receptors, Type-I (ALK3 (Activin Receptor-Like Kinase-3) and ALK6 (Activin Receptor-Like Kinase-6)) and Type-II, BMPRII (Bone Morphogenetic Protein Receptor Type-II), respectively, and form a heteromeric signaling complex acting in series. In the presence of ligand, the Type-II receptor phosphorylates the Type-I receptors, which activate signaling by intracellular effectors SMADs (SMAD (Sma and MAD (Mothers Against Decapentaplegic) Related Proteins) and TAK1 (Transforming Growth Factor-Beta-activated Kinase-1). Among the members of SMADs, SMAD1 (SMAD (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-1), SMAD5 (SMAD (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-5) and SMAD8 (SMAD (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-8) transduce signals from BMPs specifically, while SMAD4 (SMAD (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-4) is a general partner of ligand-specific SMADs. Co-overexpression of SMAD1 and SMAD4 induce differentiation of cardiac precursor cells into Cardiomyocytes and overexpression of SMAD6 (SMAD (Sma and MAD (Mothers Against Decapentaplegic) Related Protein-6), an inhibitory SMAD blocks the signal transduction and inhibits differentiation of the Cardiac precursor cells into Cardiomyocytes. BMP pathway, on the other hand, is negatively regulated by Noggin. Noggin binds specifically to BMP2 and BMP4 with high affinity and also to BMP7 with lower affinity, thereby abolishing the activity of BMPs by blocking the binding of BMPs to cognate cell surface receptors (Ref.3, 4 & 5).
TAK1 and the SMAD pathways, activated by BMPs, act in parallel to promote Cardiogenesis by activating several transcription factors like ATF2 (Activating Transcription Factor-2), NKX2.5 and GATA4. ATF2 is critical for Cardiomyocyte differentiation. ATF2 stimulates the Beta-MHC (Beta-Myosin Heavy Chain) promoter activity synergistically with SMAD1, SMAD4 and TAK1 and promotes terminal Cardiomyocyte differentiation. ATF2-induced transactivation of Beta-MHC gene depends on both the SMAD and the TAK1 pathways. Single amino acid changes in Beta-MHC result in abnormal actomyosin interactions, confirming the primary role of missense mutations in Beta-MHC gene in the etiology of hypertrophic Cardiomyopathy. Apart from stimulating Beta-MHC, ATF2 also plays a pivotal role in transactivation of some Cardiac-specific genes like NPPA (Natriuretic Peptide Precursor-A) and NPPB (Natriuretic Peptide Precursor-B). The hetero-oligomer of SMAD3 and SMAD4 bind directly to ATF2 through the MH1 (MAD Homology-1) region of SMAD3 and SMAD4 and the bZIP region of ATF2 and enhance the transactivating capacity of ATF2. NPPB is the major heart secretory product that is also accepted as clinical markers of the diseased Heart and it is also controlled by GATA factors. Other cardiac transcription factors like NKX2.5 (NK2 Transcription Factor Related Locus-5) and GATA4 (GATA Binding Protein-4) also plays pivotal role in the Cardiogenic BMP signaling pathway. Transactivation of NKX2.5 and GATA4 is mediated by TAK1. Subsequently, NKX2.5 and GATA4 induce differentiation into Cardiomyocytes cooperatively with unknown factor(s) induced by DMSO (Dimethyl sulfoxide). Although some cardiac-specific genes, such as MEF2C (MADS Box Transcription Enhancer Factor-2 Polypeptide-C) and MLC2v (Ventricular Myosin Light Chain Type-2), are upregulated by NKX2.5 and GATA4 alone, differentiation into beating cardiomyocytes requires the cooperative effects of both NKX2.5 and GATA4. MLC2v and MEF2C are positively regulated by NKX2.5. MEF2C binds to the AT-rich element in regulatory regions of numerous muscle-specific genes. GATA4 binds to the WGATAR motif in promoter regions of Cardiac- or Gut-specific genes. Both MEF2C and GATA4 are expressed simultaneously in the precardiac mesoderm along with NKX2.5 and plays an important role in cardiac differentiation. Other members of the TGF-Beta (Transforming Growth Factor-Beta) Superfamily have also been involved in induction of the mesoderm in Xenopus, Zebrafish, Chicken and Mouse. TGF-Beta is expressed early in the Cardiac region of the mesoderm (Ref.6 & 7).
Besides BMPs, another protein involved in the induction of Cardiogenesis includes Cripto. Cripto is the original member of a family of vertebrate signaling molecules, the EGF-CFC family which includes: Human, Mouse and Chick Cripto; Human and mouse Cryptic, Xenopus FRL1 and Zebrafish one-eyed pinhead. All EGF-CFC proteins contain a signal sequence for extracellular secretion, a characteristic EGF-like domain, a second cysteine-rich region called the CFC domain, and a hydrophobic C-terminus. Nodal signaling is required to support Cripto-regulated Cardiac induction and differentiation in ESCs (Embryonic Stem Cells). Cripto interacts with ALK4 (Activin Receptor-Like Kinase-4) to permit Nodal binding to the ALK4/ Act-RII (Activin type II serine/threonine kinase Receptor) complex, leading to SMAD phosphorylation. In addition, Cripto is also implicated in Nodal signaling via the orphan receptor ALK7 (Activin Receptor-Like Kinase-7), since its expression enhances the ability of ALK7 and ActRIIB to respond to Nodal ligands. Membrane-bound Cripto recruit Nodal to an Activin type I receptor (ALK4 or ALK7) and to ActRII receptors; upon receptor activation, intracellular effectors SMAD2 and/or SMAD3 are phosphorylated and accumulate together with SMAD4 in the nucleus. The intracellular mediators, in combination with co-factors then switch on transcription of Cardiac specific genes. Addition of Nodal-antagonist Cer1 (Cerberus Short), which specifically blocks Nodal by direct binding to the ligand, results in a strong inhibition of Cripto activity in promoting Cardiogenesis and provides direct evidence for a functional role of ALK4/Nodal pathway in Cripto-mediated specification of the Cardiac lineage (Ref.8, 9 & 10).
Proteins of the WNT family are also known regulators of Cardiomyogenesis. WNTs can bind to Fz (Frizzled) receptors on target cells to activate different signaling pathways. Activation of the canonical pathway leads to stabilization of Ctnn-Beta (Catenin-Beta) through inactivation of GSK3Beta (Glycogen Synthase Kinase-3-Beta), to Ctnn-Beta -dependent activation of TCF (T Cell Factor)/ LEF (Lymphoid Enhancer Factor) transcription factors and induction of WNT-responsive genes. In contrast, WNT11 (Wingless-related MMTV integration site-11) signals through a Ctnn-Beta -independent non-canonical pathway involving PKC (Protein Kinase-C) and JNK (Jun-N-terminal kinase). WNTs induced Myogenic specification and mammalian Cardiac myogenesis. WNT3A (Wingless-related MMTV integration site-3A) upregulated early Cardiac markers in Mouse through Ctnn-Beta. Wnt11-induced Cardiac differentiation in Xenopus and in Murine Embryonic Cell. TAK1 and NLK (Nemo-Like Kinase) add to the list of components that can mediate a WNT signal. NLK can antagonize Ctnn-Beta dependent signaling, as well as act as a co-activator of this pathway. Different WNT signaling pathways act as an intertwined and partially cross-regulatory network. The secretion of WNT inhibitors (such as Cerberus, Dickkopf and Crescent) by the anterior endoderm prevents WNT3A and WNT8 (Wingless-related MMTV integration site-8) secreted by the neural tube from inhibiting Heart formation (Ref.11 & 12).
In Mouse, at least two lines of signaling act together to induce Cardiac differentiation: first, a Ca2+-dependent activation of Ca2+/ Calmodulin-dependent kinase, and second, a NOX4 (NADPH Oxidase-4)-dependent activation of p38 MAPK (Mitogen-Activated Protein Kinase) through a moderate increase in ROS (Reactive Oxygen Species). These two signaling pathways converge at the level of MEF2C whose nuclear translocation requires the activation of both pathways. Although ROS play a critical role as intracellular signaling molecules, the molecular targets of ROS in Cardiomyocytes remain unclear, in particular during Cardiogenesis. Recently, it has been reported that NADPH-dependent ROS can also play a role in ESC during Cardiotrophin-1–induced Cardiac proliferation. NOX4-generated ROS leads to p38 phosphorylation and the subsequent MEF2C nuclear translocation. This crucial transcription factor is responsible for the activation of several Cardiac-specific embryonic genes, including MLC2v. MEF2C regulates the activity of MLC2v promoter. MEF2C is necessary for Cardiac phenotype determination, transcription of sarcomeric proteins, and myofibrillogenesis (Ref.1 & 13).
In addition to other transcription factors like GATAs, NKX2.5 and MEF2C, Tbx5 and Tbx20 also play an important role in Cardiac differentiation pathway. Tbx5 is a member of the T-box family of transcription factors, a family of proteins that are required for normal vertebrate patterning and differentiation. Human Tbx5 plays an important role in Heart development. Tbx5 is localized to the nucleus, it binds to DNA in a sequence specific fashion and it regulates the transcriptional level of its target genes. At least one of the targets of Tbx5 either directly or indirectly functions to control the progression of the embryonic cardiac cell cycle. Tbx5 could function to induce the expression of a Growth factor, for example EGF (Epidermal Growth Factor) or FGF (Fibroblast Growth Factor), which in turn is required for cell cycle progression. In the absence of this Growth factor the cell cycle may not proceed through to the completion of G1. Alternatively, Tbx5 also functions to regulate the expression of a key component of the pre-replication complex. In the absence of this key component the pre-replication complex may not assemble or may not load onto the ORCs (Origins of Replication), thus blocking DNA synthesis and hence cell cycle progression. Cell cycle progression through G1/S is regulated by members of the E2F (E2F Transcription Factor) family of transcription factors, the function of which is governed through their interaction with the Rb (Retinoblastoma) protein. In its hypophosphorylated state, Rb interacts with E2F, inhibiting its transcriptional activation activity. Upon Rb phosphorylation, the Rb-E2F interaction is disrupted and E2F is released and able to activate its downstream genes required for S-Phase entry. Thus, one function of Tbx5 may be to indirectly regulate the state of Rb phosphorylation. Finally, Tbx5 may function to negatively regulate general cell cycle inhibitors such as p27Xic in Xenopus. Tbx5 depletion leads to a G1/S-phase arrest, which causes a dramatic increase in the expression of proteins associated with the Cardiac cell cycle S-phase, including CDC6 (CDC6 Cell Division Cycle-6), CcnE2 (Cyclin E2), SLBP (Stem-Loop (histone) Binding Protein) and PCNA (Proliferating Cell Nuclear Antigen). The initiation and the maintenance of a Cardiac program is the result of temporally and spatially well-orchestrated interactions between all the above factors and analyses of these interactions in animal models will be very helpful for the progress of cardiac stem cell research (Ref.14 & 15) .
References
- 1
- Moorman AF, Soufan AT, Hagoort J, de Boer PA, Christoffels VM. Development of the building plan of the heart.
- 2
- Pandur P. What does it take to make a heart?
- 3
- Angello JC, Kaestner S, Welikson RE, Buskin JN, Hauschka SD. BMP induction of cardiogenesis in P19 cells requires prior cell-cell interaction(s).
- 4
- Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S, Shimazaki T, Ogawa S, Okano H, Fukuda K. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells.
- 5
- Qi X, Yang G, Yang L, Lan Y, Weng T, Wang J, Wu Z, Xu J, Gao X, Yang X. Essential role of Smad4 in maintaining cardiomyocyte proliferation during murine embryonic heart development.
- 6
- Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation.
- 7
- Lim JY, Kim WH, Kim J, Park SI. Induction of Id2 expression by cardiac transcription factors GATA4 and Nkx2.5.
- 8
- Anton R, Kuhl M, Pandur P. A molecular signature for the "master" heart cell.
- 9
- Barron M, Gao M, Lough J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative.
- 10
- Foley AC, Korol O, Timmer AM, Mercola M. Multiple functions of Cerberus cooperate to induce heart downstream of Nodal.
 关于我们
关于我们